Introducing Vanderbilt MyColony
This initiative is endorsed by the Vanderbilt Institutional Animal Care and Use Committee.

Today, we are happy to extend an invitation for you to join MyColony. Transnetyx and Vanderbilt are excited to partner together to ensure reproducible research.
For your participation, you will receive:
-
Fast, Guaranteed Accurate, Automated Genotyping
-
Institutional TAGCenters
- Free Artificial Mouse Intelligence
-
Transnetyx Automated Genotyping
-
Step 1: Automated Genotyping
-
Trasnsnetyx Colony
-
Step 2: Colony
Fast, Easy, Accurate
Eliminate human error and accelerate your research by outsourcing genotyping. Transnetyx delivers 99.97% accuracy with online results in 24 to 72 hours. Follow the link above for the first step to get started in MyColony, which is to register for Transnetyx Automated Genotyping.
Step 1: Register for your Genotyping Account
- To get started, please fill out this intake form
- Once you complete the intake form, please register here
- Upon registration, please enter the name of your PI under "Lab Name"
Accurate and efficient colony management
Create greater efficiencies by accessing, updating, and sharing your data in real time wherever and whenever. With streamlined communication and convenient task management, Transnetyx Colony software promotes efficient practices and reduces the number of non-scientifically useful animals. Follow the above link for the second step of MyColony.
Step 2: Get Started in Colony
-
All labs must complete genotyping assay development and validation before starting Colony training
- Do not operate in the Sandbox. The Sandbox is for learning purposes only.
- In Colony, start by entering a VUMC/VU PI Name as the Group Name. This will allow the PI to see all records while also allowing the personnel associated with the PI (e.g. staff, Co-PIs, Postdocs, sturents) to enter and track populations separately.
-
To access Colony and get started, please access our Support Hub here
-
Labs will receive personalized training from the Colony by Transnetyx team, prioritized by order of census size
Testimonials
I realized I have been setting up the crosses a lot faster, because I got the genotyping results a lot faster. Everything was in the system, so I knew exactly which mice to pull and which to cull. The Colony system made it very easy and it just became second nature. When you multiply this by strain, it adds up very quickly.
We, meaning the entire med school research community, have run out of space to house animals. With lots of research dollars coming in to continue our work but with no space to expand, we needed a better way to manage our colony. Your software and the integration with the genotyping service was the answer.
MyColony Newsletter
If you missed the previous newsletters, follow the links below to learn more!
Q3 2024 Newsletter | 2024 March Newsletter | 2023 December Newsletter | 2023 August Newsletter | 2023 September Newsletter
Frequently Asked Questions
Who owns the data in Quick Order (Assay Development) and Colony (Genotyping orders, results, and colony management)?
You own the data!
If Transnetyx goes out of business, will I be able to retrieve my data?
Yes. All data is backed-up and will be provided to all researchers that we serve.
Is the software as easy as my Excel spreadsheet?
Colony is different from Excel but not more difficult than Excel. Excel is a spreadsheet – Colony is a tool. Options in Colony far exceed the capabilities that Excel provides. Please know that Colony has two main views: List View and Cage Map View. Starting in List View may feel more comfortable because it mimics basic Excel views. As you advance in Colony, you’ll naturally become fonder of advanced features like the Map View.
Is the software user friendly, easily accessible, intuitive, and flexible enough to conform to individual lab nomenclature and notes?
User Friendly: YES
Easily Accessible: YES
Intuitive: YES
Flexible: YES
Researchers have no limits to the number of people that can be added to a GROUP (lab account) in Colony. Processes can be set to fit your lab’s specific needs. In Colony, you will dictate strain nomenclature and can input notes, task reminders, animal use cage (ex: experimental), treatments, and more.
Are there other companies that offer similar services and, if so, have we explored partnering with them?
There are other companies that offer genotyping services; however, not fully automated, guaranteed accurate genotyping services with a guaranteed 72-hour turn around time. Transnetyx is the only laboratory that does not require human hands – preventing the threat of human contamination. Transnetyx is the only company providing free assay development for real-time (qPCR with TAQman probe) assays. Furthermore, Transnetyx is the only company that offers genotyping and genetic monitoring using the same biopsy. Vanderbilt only agreed to partner with Transnetyx after confirming their industry reputation and recognized performance.
Where is Transnetyx located?
Transnetyx is located in Memphis, TN where it processes an average of more than 100,000 samples/week from leading research institutions all over the globe.
Can I change my plan later?
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur.
What happens if Transnetyx makes a mistake?
Transnetyx guarantees accurate results in 72-hours or less. IF there are results concerns, Transnetyx will immediately retest samples at no cost to the lab. If required, Transnetyx will redesign/optimize the initial assay designed. Vanderbilt will not be charged until accurate results are produced. In short, Transnetyx will be accountable for errors resulting from assay development and/or results review.
How stable is Transnetyx?
Transnetyx is a very stable company established in 2004 that has processed over 40 million samples globally. Transnetyx processed over 5 million samples in 2023, representative of over 10% growth in sample processing since 2022.
In addition to their core Genotyping and Colony offering, Transnetyx provides:
What happens if Transnetyx goes out of business?
Worst case scenario is that labs go back to performing their own genotyping and Transnetyx contract costs are removed from the annual per diem rate calculations. You will still have lifetime access to your data.
Is there a financial relationship between VUMC or VU and Transnetyx?
Outside of the agreement to help Vanderbilt to serve your research, there is not a financial relationship between VUMC or VU and Transnetyx.
Will Transnetyx design probes for mutations that have not been published in literature?
Transnetyx will genotype all lines except founder lines. Transnetyx can genotype F1 generation models if the correct mutation has been confirmed in the founders. Please note that Transnetyx is doing beta-testing with Vanderbilt’s VGER in this area.
Can I get my tissue (tail, ear punch, liquid DNA, etc.) back?
Transnetyx first lyses the tissue received and purifies the DNA. 1/3 of the lysate is utilized for genotyping and the other 2/3 is stored for up to four months. When testing a strain for the first time, Transnetyx provides five free samples for any strain that has been custom designed. This is an excellent opportunity to validate the assay(s) put in place as opposed to having to manually genotype side-by-side.
Researchers can request the lysate to be sent back; however, routine/repeated requests will incur a charge.
How long does Transnetyx keep my tissue?
Transnetyx stores tissue for up to four months.
Sometimes I need to retest – how does that work?
You can request retests online.
Can I change my plan later?
Lorem ipsum dolor sit amet, consectetur adipiscing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur.
If there is a results discrepancy, do I have to pay for a retest?
No.
What is the average actual turnaround time?
Transnetyx guarantees accurate results in 72-hours or less; however, average completion is between 48-56 hours.
How does Transnetyx use qPCR to determine homozygosity?
At Transnetyx, we exclusively operate a high-throughput, real-time PCR-based system for genotyping. Our TaqMan assays target specific sets of DNA sequence in either the targeted or wildtype alleles. This means that zygosity for a locus is not determined through quantitative analysis, but by testing for each potential allele that could be present at that locus.
This means that randomly integrated transgenes (without a known integration site) will not have zygosity reported, as a single presence/absence assay will be run. We can provide you with the raw data for these transgenes which normalizes the probe signal back to a single copy housekeeping gene. You may use this for your own interpretation of copy number; however, we cannot guarantee that this raw data will be able to provide you copy number.
Are mice housed at the VA eligible?
Yes.
Will there be a TAGCenter in the VA?
Researchers with animals in the VA will be provided with packaging to submit samples directly back to Transnetyx. There will not be an additional cost.
There are currently two TAGCenters at Vanderbilt University. One of the TAGCenters is located in the MCN Facility Entry - 7th Floor just outside of the animal care facility. Elevators 20 & 21 will take you directly to the unit. The shipping days for this TAGCenter are Monday by 3:00 PM and Wednesday by 3:00.
The newest TAGCenter is located in the PRB ~ Located outside the PRB animal facility to the left of Room 801. The shipping days for this unit are Tuesday by 3:00 PM and Thursday by 3:00 PM.
Is 24-hour service available?
Yes, For an additional charge.
Can I opt out?
No. Effective July 1, 2023, existing cages of animals ordered directly from a vendor and not bred for subsequent experimentation (as well as future orders of a similar nature) will be assessed the Standard Mouse Per Diem Rate. Existing and new cages of animals bred or born at Vanderbilt will be assessed the MyColony rate whether or not Transnetyx genotyping services are used. This decision was made to encourage the use of available services (automated genotyping and mouse colony management software) which have consistently been shown to reduce genotyping errors/cost, improve breeding colony productivity, and utilize limited housing space more efficiently. These outcomes are essential to improving the rigor and reproducibility of Vanderbilt research studies using mouse models and allow for more responsible use of animals and resources.
Why do I have to pay for services I don’t want to use?
Use of MyColony (automated genotyping and mouse colony management software) has consistently been shown to reduce genotyping errors/cost, improve breeding colony productivity, and utilize limited housing space more efficiently. These outcomes are essential to improving the rigor and reproducibility of Vanderbilt research studies using mouse models and allow for more responsible use of animals and resources.
What mouse per diem rate will my mouse cage receive?
My IACUC protocol includes experiments that involve breeding genetically engineered mice (GEM) and C57BL6/J control animals. Offspring may or may not be genotyped and are used in subsequent experiments. Occasionally I order mice from a vendor to refresh my lines (GEMs and controls).
- Rate charged for cages of GEM bred at Vanderbilt: MyColony Per Diem Rate
- Rate charged for cages of control animals bred at Vanderbilt: MyColony Per Diem Rate
- Rate charged for cages of mice ordered from a vendor to refresh lines: Standard Per Diem Rate for the life of the cage card. The MyColony rate will be applied to cages of offspring as pups are generated and weaned into new cages.
- Rate charged for cages of experimental mice generated from breeding conducted at Vanderbilt: MyColony Per Diem Rate
My IACUC protocol includes experiments that involve ordering animals directly from a vendor and using them in “all-in/all-out” experiments (e.g. nude mice ordered in, injected with tumor cell line, and euthanized after 6 months). No breeding at Vanderbilt is conducted to generate the experimental animals or as part of the experiments.
- Rate charged (throughout duration of experiment) for cages of experimental mice ordered from a vendor: Standard Per Diem Rate
My IACUC protocol includes both types of experiments described above. Will I have to pay the MyColony Rate for all my cages?
- No. The rate applied will depend on whether the cage houses animals generated (bred) at Vanderbilt or procured directly from a vendor. A single IACUC protocol may be charged the two different rates. The cage card identifies which rate applies to which cage.
Fast, easy, and accurate Automated Genotyping.
Our Automated Genotyping service:
-
Eliminates human error.
-
Saves you time
-
Accelerates your research.
Streamlined Software
for a Simplified
Colony Management.
Poor visibility in your colony can lead to unnecessary cages, non-productive breeders, and slower research. As part of the MyColony, you have access to our FREE Colony management software designed to let you access, update, and share breeding and Colony data in real time.
More science in every cage.
Manage your colony more efficiently with less effort. Based on your research goals and resources, AMI will:
- Guide you to a more efficient cage footprint
- Reduce the number of animals housed over the course of your project
- Optimize your production of viable experimental subjects
You will have access to a free AMI trial for a year. After the free trial, AMI will cost four cents per cage per day.
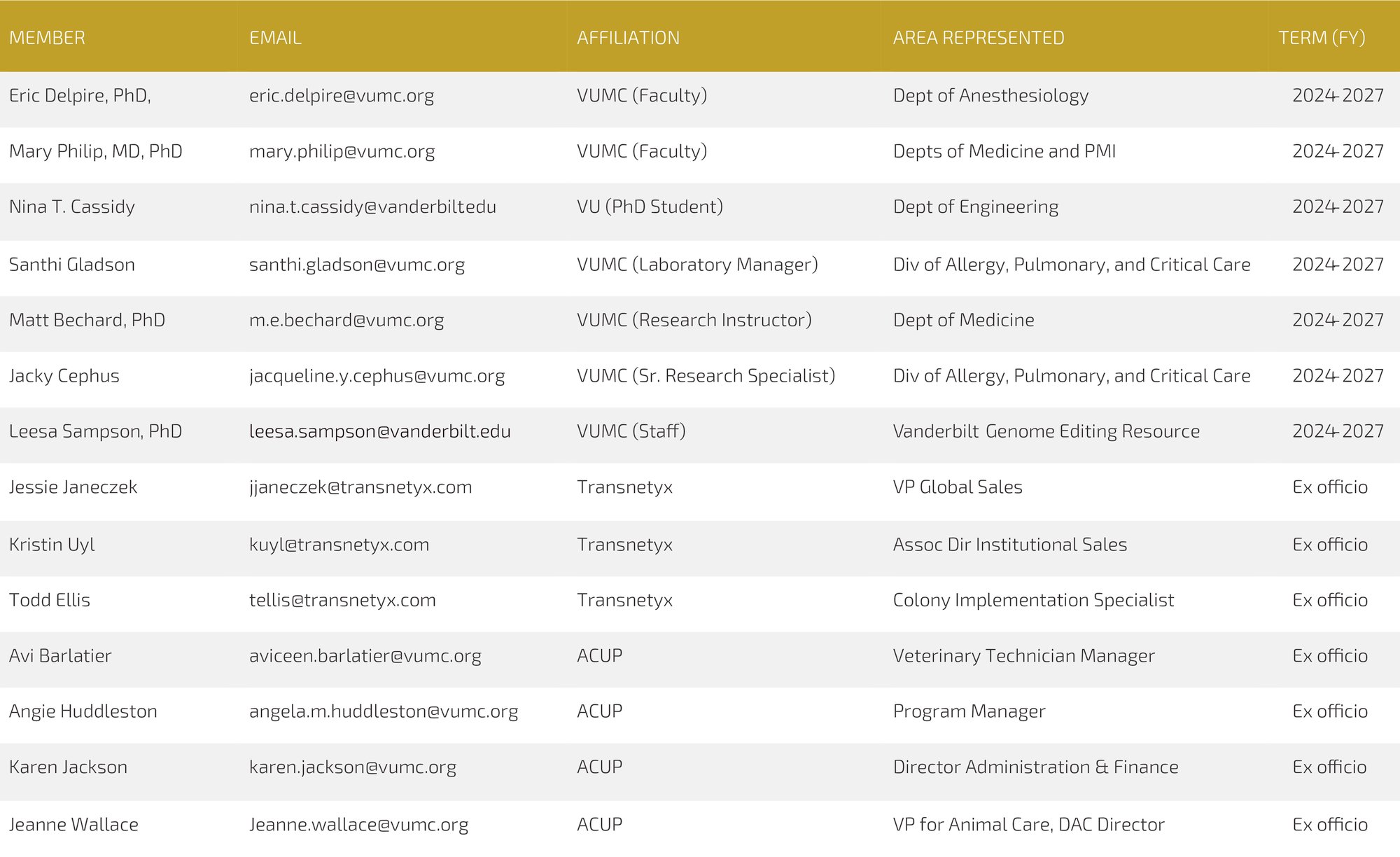
MyColony User Group
The Animal Care and Use Program (ACUP) Leadership and Transnetyx recognize the significant value of including the perspective of research faculty and staff as part of implementing and maintaining MyColony, an initiative launched in 2023 consisting of automated genotyping services and mouse colony (experimental and breeding) management software. Use of MyColony (automated genotyping and mouse colony management software) has been shown to reduce genotyping errors/cost, improve breeding colony productivity, and utilize limited housing space more efficiently. These outcomes are essential to improving the rigor and reproducibility of Vanderbilt research studies using mouse models and allow for more responsible use of animals and resources.
The MyColony User Group has been established to create open forums for discussion between ACUP Leadership, Transnetyx representatives, and the Vanderbilt research community.
Goals
-
-
Advise ACUP and Transnetyx representatives on process/software improvements, as well as future use/integration designed to enhance Vanderbilt research resources
- Identify and prioritize action items that will maximize the value and support adoption of MyColony services and software by Vanderbilt faculty/staff
- Assist with development of resources/activities aimed at facilitating adoption of MyColony services and software
- Serve as a resource on the specific features of MyColony of highest value to other Vanderbilt research faculty/staff
-